Critical Assessment of protein Structure Prediction, or also known as CASP, is a where researchers worldwide attend to showcase their experiments on protein structure prediction.
The once every two years event (since 1994), provides researchers the opportunity to objectively test their prediction methods, and deliver their independent assessment of the art in protein structure modeling to the research community and software users.
The goal is to help the global advancements in methods for identifying protein three-dimensional structure from its amino acid sequence, considered as one of the biggest puzzles in biochemistry.
Many researchers in this field of science view the experiment as a “world championship", with more than 100 research groups from all over the world participate in CASP on a regular basis.
And here, Google DeepMind has beaten all of those researchers at CASP, by a huge margin.

DeepMind manages to accomplish such feat using machine learning technology. What this advancement represents fascinate both biochemistry and AI in general.
According to a DeepMind's blog post:
The AI is called AlphaFold, and has been in the works since 2017.
Proteins are large, complex biomolecules essential in every living thing, and affects nearly every function a body's perform.
From muscle contraction to sensing light, and even turning food to energy, proteins can be traced back to even more proteins. What makes those proteins, is called genes, and is encoded in the organism's DNA.
And what any given protein can do, depends on its unique 3D structure.
But figuring out the 3D shape of a protein just from its genetic sequence is a complex task that scientists have found challenging for decades. Predicting how those amino acid residues inside proteins is known as “protein folding problem”.
Over the decades, researchers have been able to determine the shapes of protein inside labs using experimental techniques. But each method involves a lot of trials and errors.
This in turn makes the research years to complete, with cost hovering at tens of thousands of dollars per structure.
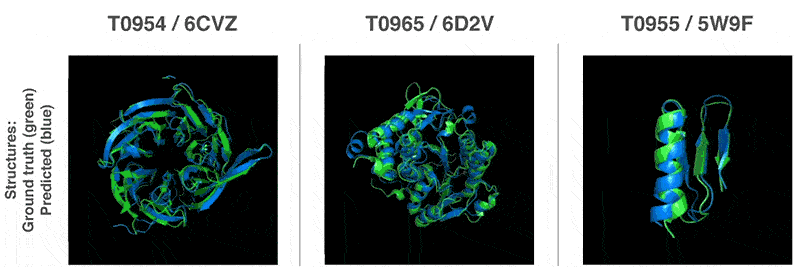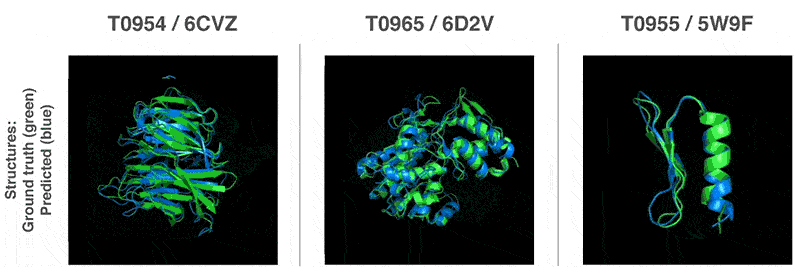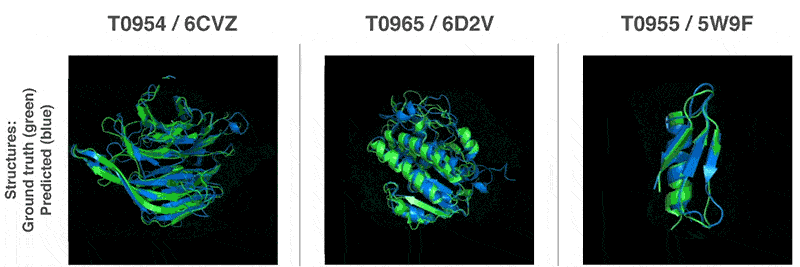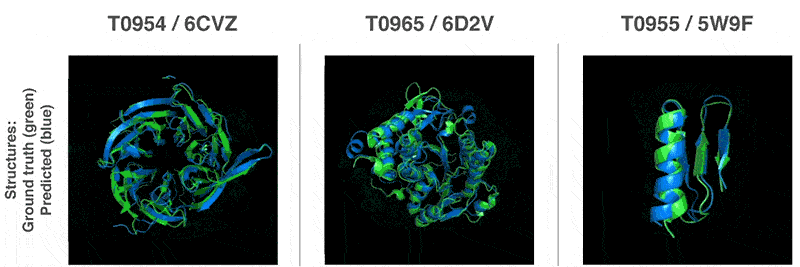
This is why biologists are using AIs as alternatives to this process.
With the advancements of other technologies in biology, researchers have reduced the cost of genetic sequencing, thus providing a lot of data for Google DeepMind's AlphaFold to learn from.
"DeepMind’s work on this problem resulted in AlphaFold, which we submitted to CASP this year," said DeepMind.
"We’re proud to be part of what the CASP organisers have called 'unprecedented progress in the ability of computational methods to predict protein structure,' placing first in rankings among the teams that entered (our entry is A7D)."
"The success of our first foray into protein folding is indicative of how machine learning systems can integrate diverse sources of information to help scientists come up with creative solutions to complex problems at speed."
There is still a lot of work to be done before the AI can have a quantifiable impact on treating diseases, managing the environment, and more.
But here, the potential is indeed enormous. DeepMind has already assembled a dedicated team focused on delving into how machine learning can advance the world of science.
